自20世纪发现青霉素以来,抗生素被广泛用于治疗和预防细菌感染,并作为饲料添加剂大量投入到畜牧和水产养殖业中[1-2]。伴随抗生素的过度使用,刺激了人体和动物肠道内抗生素抗性细菌(antibiotic resistant bacteria, ARB)与抗生素抗性基因(antibiotic resistance genes, ARGs)大量产生[3]。畜禽粪便携带ARB和ARGs被动物排出体外,作为有机肥料进入农田环境。我国畜禽粪便年产量约38亿t[4],粪肥还田利用价值巨大。畜禽粪便是ARGs的重要储存库,长期施用促进了有机肥源ARGs在土壤中增殖扩散,并通过水平基因转移(horizontal gene transfer, HGT)进入土壤微生物。植物是污染物从土壤环境进入人体的桥梁,与人类健康息息相关,土壤-植物系统也被视为地球生态系统中同人类生存和健康最密切的部分,因此关注ARGs在土壤-植物系统中的传播过程与影响机制具有重要意义。
土壤-植物系统包括植物及其根系周围的土壤环境,ARGs可以伴随“土壤-根系”或“土壤-空气-叶际”中微生物的转移进入植物,通过食物链传递给人体。土壤中的抗生素抗性以及植物获得ARB和ARGs过程共同决定了土壤-植物系统中抗生素抗性的传播。部分土著微生物本身具有一定的抗生素抗性[5],人类活动促进了土壤中抗生素抗性的传播[6-8],土壤动物也是ARGs的重要携带者[9-11],以上共同作用使得土壤成为ARGs的重要源和汇。一些耐药菌将ARGs水平转移到土壤微生物再进入植物体内,或通过根系直接进入植物微生物组,在植物微生物组中继续垂直或水平传递[12]。在此过程中,根系分泌物、根系结构与形态以及根际环境基因水平转移频率对ARB和ARGs从根际向植物的迁移发挥重要作用[13]。除“土壤-根系”外,“土壤-空气-叶际”也是土壤抗性组进入植物的重要路径。翻耕和土壤扬尘会造成土壤微生物沉降到植物地上部,部分微生物在叶际定殖或通过叶表气孔、组织伤口进入植物体内[14]。目前关于ARGs和ARB如何从叶际进入植物内部的过程与机制尚不明确。
厘清土壤-植物系统中影响ARGs的重要因素,可以为阻断ARGs传播提供思路。除土壤与植物本身,土壤-植物系统中其他因素也影响了ARGs的传播。根瘤菌在豆科植物中广泛存在,通过固定空气中的游离氮素提供给共生植物,所形成的根瘤结构具有丰富的ARGs,并促进了ARGs的水平转移[15]。真菌菌丝以及真菌与植物根共生形成的菌根不仅增加了细胞间接触频率[16-17],为ARGs的水平转移提供便利,还为ARB从土壤向植物转移提供介质。此外,污染物、环境胁迫、土地利用方式与气候条件也会改变土壤-植物中抗生素抗性组成。降低土壤中ARGs丰度、阻断其传播路径是控制抗生素抗性向植物传播的关键。近年来,广泛用于阻控抗生素抗性传播的主要方法有物理化学和生物控制技术,如好氧堆肥、添加土壤改良剂、动植物修复和噬菌体技术[18-21]等。以上技术单一或联合应用,有望降低ARGs通过蔬菜作物进入人体的环境健康风险。
从“大健康(One Health)”理念出发,“土壤-植物-人群(动物)”连续体中的土壤健康和微生物循环是实现“大健康”的核心[22]。食物链传递是将环境安全与人体健康建立紧密联系的渠道,而植物是污染物、耐药菌从土壤环境向人类传播的重要桥梁。土壤环境中ARGs组成复杂,含有大量病原菌,ARGs可以在同种或者不同种菌群之间传播,使得耐药机制更加复杂,一旦通过食物链被人体摄入,将增加疾病治疗难度和治疗成本[23]。因此,本文首先总结了土壤中抗生素抗性的来源与演变机制,强调了抗生素抗性从土壤向植物的传播过程与影响因素,最后讨论了调控ARGs在土壤-植物系统中传递的阻断技术,并对未来研究进行了展望。
1 土壤中的抗生素抗性(Antibiotic resistance in soil)
1.1 土壤微生物是ARGs的重要储存库
临床常用的抗生素多数来源于土壤细菌和真菌[24],如青霉素[25]、链霉素[26]和利福霉素[27]等。虽然土壤环境中天然抗生素含量很低,但仍可以对微生物产生一定的选择压力,赋予微生物抗性,从而使ARGs自然存在于土壤微生物中[28](图1)。van Goethem等[29]在未受干扰的南极表层土中发现的ARGs包含23个ARG家族,涵盖了所有已知的常见抗生素耐药机制。Galán等[30]对30 000年前永久冻土分析发现,冻土DNA中ARGs组成丰富,部分ARGs具有多重耐药性。这表明ARGs天然存在于土壤微生物中[31]。值得注意的是,土壤病毒组是土壤微生物组的重要组成部分[32],噬菌体是土壤中含量最丰富的病毒群以及ARGs传播的重要载体,在各种环境条件下对灭活和消毒剂的持久耐受性更强[33-36]。由于噬菌体的遗传物质被噬菌体衣壳蛋白包裹,噬菌体携带的ARGs可能比环境中的胞外ARGs[37-38]更稳定。噬菌体作为侵袭细菌的病毒,可以携带ARGs在土壤细菌中定殖,促进ARGs在土壤微生物组中的传播。病毒还可以通过水平基因转移,从宿主获取基因,再传递给其他宿主,促进ARGs在病毒与宿主间的传播[39]。目前,在农田土壤和新鲜蔬菜中均有携带ARGs的噬菌体被检出[40],增加了ARGs从土壤环境进入人体的传播风险。
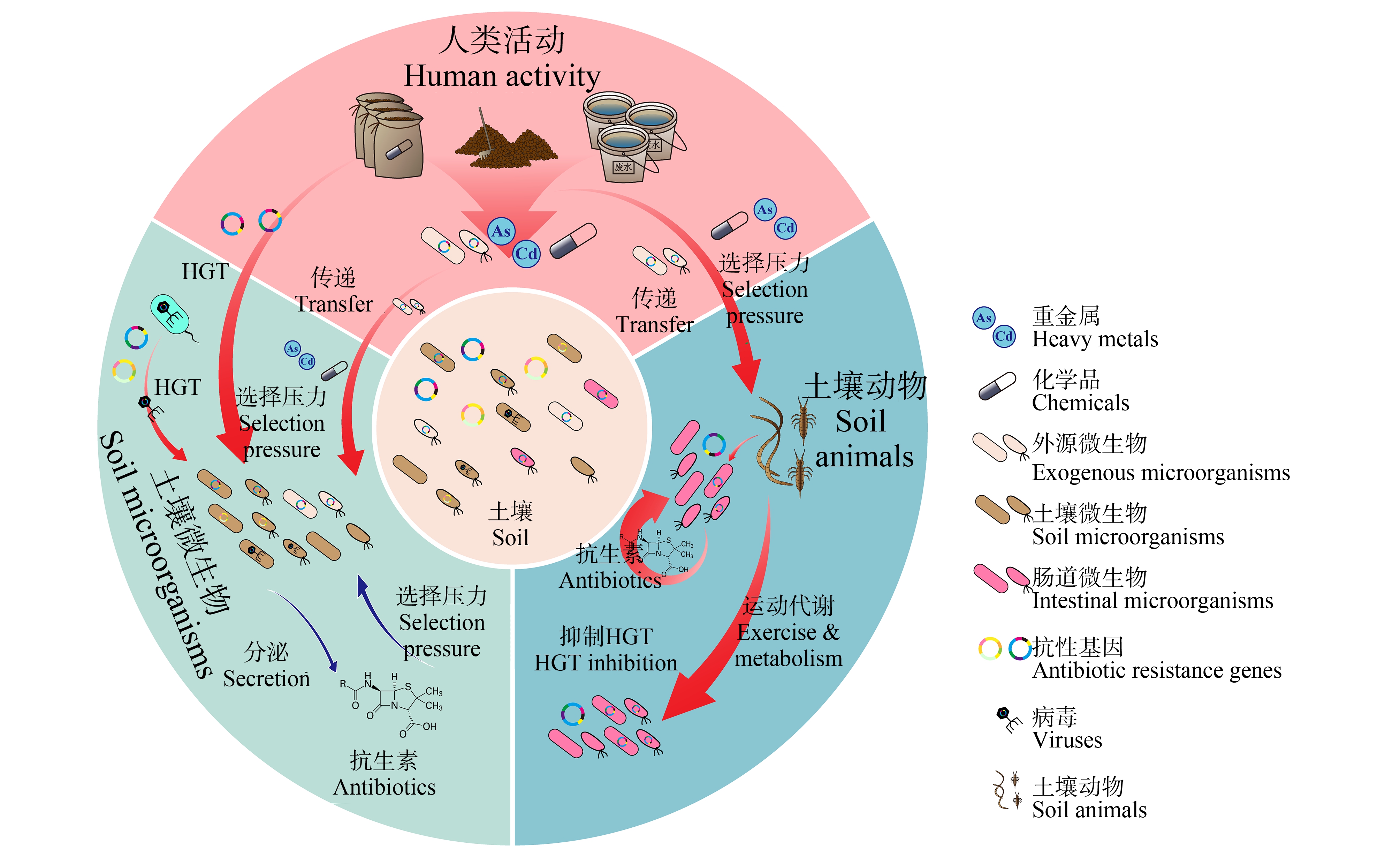
图1 土壤中抗生素抗性
注:HGT表示水平基因转移。
Fig. 1 Antibiotic resistance in the soil
Note: HGT represents horizontal gene transfer.
1.2 人类活动促进土壤中抗生素抗性传播
施用养殖粪污、污水污泥等会导致化学品残留、ARB和ARGs进入土壤环境,并通过水平转移将抗性传播到土著菌在内的其他微生物中,导致土壤微生物抗性越来越依赖于人类活动[41](图1)。畜禽粪便是ARGs的重要储存库,有机肥施用会导致ARGs和ARB从粪便进入土壤,显著增加土壤中ARGs水平[42-43]。Liu等[44]对连续3年施用有机肥的稻田分析后发现,动物粪肥的施用主要通过增加对土壤中原有ARGs的选择压力以及引入外源性ARGs来促进土壤中抗生素抗性传播,而可移动基因元件(mobile genetic elements, MGEs)和微生物群落是塑造土壤ARGs组成的关键因素。除有机肥外,由于活性污泥中含有丰富的ARGs残留,且污水处理过程中ARGs很难被彻底去除[45],因此污水污泥也是环境中ARGs的重要来源。已有研究在中国11家污水处理厂的污泥和出水中检测到200多种ARGs[46],在美国、印度和希腊的多个污水处理厂污泥和出水中也检测到了多种ARGs存在[47-49]。处理后的污水污泥可通过再生水灌溉和污泥施用方式进入城市和农田土壤,其中残留的ARGs进入土壤并长时间存在。施加污水污泥6个月后,仍能在土壤中检测到来自污水污泥的ARGs(blaNPS-2和tetA)残留[50]。此外,长期施用污水污泥会导致重金属在土壤中不断累积,如Cu和Zn等,对ARGs产生共选择压力,促进土壤中ARGs传播[51]。
1.3 土壤动物影响ARGs归趋
土壤动物是土壤ARGs的“隐藏库”,参与众多土壤生态过程,改变ARGs的传播归趋[52](图1)。土壤中的无脊椎动物肠道构成了独特的微生物群落栖息地[53],肠道微生物可以通过分泌抗生素等杀菌剂抵抗外源微生物入侵[54]。Agamennone等[55]从跳虫肠道细菌中提取了一系列抗生素,这些抗生素会诱导自身肠道微生物产生抗性,还会随代谢进入土壤环境,影响其他土壤微生物的抗生素抗性。土壤动物活动频繁,能够通过运动、取食和代谢等方式加速ARGs在土壤中的传播[9-10]。长期施用有机肥和污泥显著促进了土壤动物肠道内ARGs丰度,以及ARGs在土壤食物链中的传递[32, 56-57]。研究证明,蚯蚓可以显著降低土壤中ARGs的种类和丰度[58],提高非耐药菌丰度、促进微生物多样性[58]。不仅如此,蚯蚓能在一定程度上减少微生物组中MGEs的种类和丰度,这有助于抑制土壤中ARGs的水平传递[58]。Zheng等[59]发现ARGs可能在土壤-线虫-蚯蚓食物链中传递,Xiao等[60]发现,蚯蚓的使用降低了根际土壤ARGs的绝对丰度,并且蚯蚓可以通过食物链不断降低土壤和线虫中ARGs的丰度。因此,土壤动物不但是土壤中ARGs的重要来源,其行为活动还会改变ARGs在土壤中的行为归趋。蚯蚓是构成土壤动物生物总量的主体,基于蚯蚓的生物修复可能是降低土壤中ARGs传播风险的有效方法[61]。
2 抗生素抗性从土壤向植物的传播过程(The transmission process of antibiotic resistance from soil to plant)
2.1 “土壤-根系”是ARGs进入植物的主要途径
根际是土壤-根系-微生物相互作用的微区,是土壤微生物进入植物的主要场所(图2)。Wang等[62]通过扩增子和宏基因组测序技术分析了红树林土壤-根系的抗性组谱,提出将“土壤-根系连续体”作为一个相互连接的汇,通过该汇某些ARGs从土壤流入植物,造成ARGs在植物微生物组中进一步传播。ARGs从土壤-根系流入植物的具体途径包括:抗性微生物从根部伤口进入植物;ARGs通过水平转移进入根际微生物,再进入植物根系;土壤动物在根际活动过程中促进ARGs从根部进入植物(图2(b))。Xu等[12]利用荧光观察技术,发现携带RP4抗性质粒的大肠杆菌可以从根系进入拟南芥,并迁移到植物地上部,同时,携带抗性的RP4质粒可以水平转移到土壤微生物中再进入植物内生菌。线虫对根系的取食为土壤微生物进入植物开辟通道[63],并可在取食过程中将自身携带的ARGs传递给植物,促进抗生素抗性向植物的传播。

图2 土壤-植物系统中抗性基因的传播过程与影响因素
Fig. 2 The transmission and influencing factors of antibiotic resistance genes in soil-plant system
2.2 根系分泌物决定了根际微生物组和抗性组组成
根系分泌物决定了根际微生物的种类和活性[64],影响根际中ARGs的组成。植物根系通过释放有机酸、糖和次生代谢物等吸引某些携带ARGs的功能菌在根系附近定殖[65],被招募的微生物往往能够很好地适应植物根际的生存环境(图2(c))[66-67]。例如,小麦经过多代栽培后,植物功能菌在根际明显富集,并促进了根际环境中ARGs的富集,富集的微生物组和抗性组与调控自诱导剂、氨基酸代谢和碳水化合物代谢的基因具有显著关联性,说明某些功能基因与ARGs在根际功能微生物中存在共表达[68]。Brandt等[69]发现,暴露于磺胺嘧啶的土壤细菌,根系分泌物显著提高了细菌的抗生素抗性。植物根系也会分泌杀害或抑制病原微生物生长的活性物质,如茴香所产生的根系分泌物能够招募烟草黑胫病菌的游动孢子,根系分泌物中的4-乙基苯乙酮、香草素和N-甲酰基哌啶抑制烟草黑胫病菌生长[70],进而降低了根际环境中病原菌的侵染过程以及ARGs向病原微生物的传播风险。
2.3 根系形态与结构影响ARB从土壤向植物的传播
根系形态与结构影响微生物在根系定殖,进而影响携带ARGs的微生物进入植物根系。根系越旺盛,与土壤的接触面积越多,ARB在植物表面可定殖的面积越大;根系直径越小,微生物可定殖面积越小,对于生存空间的竞争越激烈[71],可在一定程度上抵御ARB的入侵。凯氏带和木栓层构成根系质外体屏障,能有效阻止有害生物进入植物体内[72]。在植物不同生长阶段,由于质外体屏障的变化,营养与水分的运输途径不同,因此影响ARB在植物中的转移。根系质外体屏障受生物(植物激素和病原体)与非生物因素(化学污染物、干旱和低氧等)调节与诱导[73-75]。诱导加强的根系质外体屏障可以减少病原体和ARB的入侵。但目前对于质外体屏障如何影响根系吸收ARB和ARGs的过程与机制仍不明确。
2.4 根际环境中ARGs的水平转移机制
根际环境中基因的水平转移促进了ARGs在土壤-根系中传播。在根系分泌物作用下微生物大量繁殖,极易在根际形成生物膜,有助于ARB的定殖[13]。生物膜增加了细菌之间的物理接触,便于根际微区中微生物之间的物质交换和和信息流通[76],为ARGs水平转移提供便利条件[77](图2(c))。研究发现,小麦和玉米的根际微生物中,插入序列、质粒、转座子和整合子等可移动元件显著富集,驱动了ARGs在根际与植物中的水平转移[68, 78]。不同类型植物的根系分泌物在种类和含量上存在差异,影响供体菌与受体菌密度,进而改变ARGs在根际的接合转移频率[76]。西红柿根系分泌物中的有机酸能明显提高根际细菌总量[79],种植西红柿的情况下土壤中质粒的转移频率显著高于玉米和小麦[80]。豌豆根际抗性质粒的转移频率显著高于大麦根际抗性质粒的转移频率,这很可能是因为豌豆根系分泌物更多、根际的供体菌密度更高,因此促进了ARGs的水平转移[81]。
2.5 ARGs从“土壤-空气-叶际”进入植物的传播过程
土壤中ARGs还可以通过空气传播的体外途径从叶际进入植物体内。ARB在自然条件与人类活动下(扬尘、翻耕等)从土壤进入空气,再随灰尘或气溶胶降落到叶片表面,进入叶际微生物[82](图2(c))。Deng等[83]发现“土壤-空气-叶际”是土壤中ARGs向叶际转移的主要途径,覆盖地膜后叶际ARGs降低了80.7%~98.7%,其中87.6%的叶际ARGs位于细菌染色体上,说明微生物转移和基因垂直传递对ARGs在生菜叶际传播的影响较大。与生菜叶际ARGs的传播机制[83]不同,小麦叶际ARGs与MGEs显著相关[84],说明ARGs在小麦叶际的传播可能主要受基因水平转移的影响。叶际微生物可以通过侵染植物组织和气孔进入叶片内部[14]。气孔在植物叶片中普遍存在,以叶片下表皮居多,其尺寸通常在5~10 μm×20~40 μm之间,大于环境中大多数细菌尺寸,是许多病原菌的侵入通道[85],因此ARB可从气孔进入植物内部成为植物内生菌(图2(a))。气孔数量、尺寸及闭合过程会影响叶片对ARGs的吸收。此外,植物叶片的次生代谢产物(β-胡萝卜素、2-苯乙基硫代葡萄糖等)[86]和挥发性有机化合物(小分子的如:二氧化碳、丙酮、萜类、醛类和醇类等,大分子如:长链烃、倍半萜类等)[87]会影响叶际微生物的种类与活性[88]。因此我们推测,携带ARGs的微生物在叶际-叶内的传播受叶片结构与叶片分泌物的共同影响。但目前关于ARGs和ARB从叶际进入叶片内部的过程与机制尚不明确。
2.6 ARGs在植物中的转移与分布特征
ARGs在植物中的分布具有普遍性和多样性,可存在于在根、茎、叶、果实和种子等组织和器官中[89](图2(c))。同一种植物的根际微生物、内生菌以及叶际微生物抗生素抗性明显不同[90-91]。根系与土壤接触,因此根际微生物抗性主要来自于土壤微生物。特别是在施用有机肥的情况下,会将大量ARGs引入农田环境[92],促进了作物根际环境与作物地下部ARGs丰度。外源物质进入根系后,会随植物蒸腾作用向地上部迁移,因此蒸腾作用可能是造成植物体内游离ARB从地下部向地上部迁移的重要动力[13]。通常,根际内生菌中ARGs的种类和丰度要高于叶片,可能是因为ARB向上迁移,阻力不断增加,因此种类和丰度降低。例如,有机肥源ARGs从根际进入生菜内生菌[93],从地下部向地上部,有机肥源ARGs的种类不断减少。通过土壤-空气-叶际传播途径,携带ARGs的土壤颗粒直接沉降到叶片表面进入叶际微生物,避免了ARGs从植物地下部向地上部迁移的阻力,但叶际ARGs是否能向下传递进入植物根部,目前仍未见报道。内生菌在植物体内主要是以生物膜形式存在[94],因此ARB一旦形成生物膜后,其移动性也会大大降低,但细菌细胞之间接合转化以及信号传递作用增强[95],ARGs的水平转移频率提高。此外,只有部分根际或叶际微生物可以在植物体内存活成为内生菌,且不同内生菌占据不同的生态位[96],受植物内部理化环境、微生物特性等因素共同影响。因此在根系中能够生存的ARB可能在茎叶中无法存活,反之亦然。植物能通过果实和种子将内生菌垂直传递到子代[96],在马铃薯[97]、草莓[98]和生菜种子[99]中均有ARGs检出,因此ARGs也可以随果实和种子传递到子代作物中。然而,也有部分研究表明,有机肥或废水加入土壤后,虽然会极大提高土壤中的ARGs丰度,改变微生物群落结构,但对作物的果实和种子(如玉米、番茄)并没有产生显著影响[100-101]。目前,通过内生菌传递抗生素抗性到植物子代的传播规律和影响机制尚不明确。
3 抗生素抗性在土壤-植物中传播的影响因素(The influencing factors of antibiotic resistance from soil to plant)
3.1 根瘤菌促进豆科植物ARGs的传播
根瘤菌与豆科植物互利共生,是豆科植物的重要内生菌,抗生素抗性在紫花苜蓿[102]、银合欢[103]和胡芦巴[104]等豆科植物的根瘤菌中普遍存在(图2(c))。在植物根系所分泌的杀菌剂和植保素作用下,根瘤菌极有可能演变出较强的抗生素抗性,促进ARGs在根瘤菌以及其他内生菌中的传播[105]。此外,质粒能够在根瘤菌之间发生接合转移[15],豆科植物的根瘤结构提高了细菌密度[106],促进了ARGs的水平转移频率。Liu等[105]发现,豆科植物南苜蓿和紫云英的根瘤结构是ARGs存在的主要场所,根瘤菌是ARGs的主要潜在宿主,抗性质粒在根瘤中的水平转移能力高于非根际土。从砷污染的大托叶猪屎豆和黧豆根瘤中分离的大多数耐砷根瘤菌(革兰氏阴性菌)同时对β-内酰胺类抗生素具有耐药性[107]。主要因为一方面革兰氏阴性菌产生β-内酰胺酶,可以修饰和灭活β-内酰胺类抗生素[108];另一方面重金属抗性基因与β-内酰胺抗性基因可能在同一质粒上[109],因此砷污染的土壤会对重金属和抗生素抗性基因产生共选择,促进根瘤菌抗生素抗性。鉴于此,厘清根瘤菌中抗生素抗性的传播与演变规律对调控豆科植物中抗生素抗性具有重要意义。
3.2 真菌影响根际ARB的传播
真菌在环境中广泛存在,可以作为细菌运动的桥梁,促进ARB从土壤向根际迁移,进而增加植物吸收ARGs水平(图2(c))。真菌能够与细菌跨界合作,真菌及其菌丝、细菌簇和胞外多糖等胞外分泌物交织在一起,形成结构更稳定、耐受性更强的多细胞生物膜,并以更快的速度扩散[110]。土壤中真菌与ARB一旦形成生物膜,极有可能提高ARB微生物群落的稳定性和移动性。菌丝分泌物可以将微生物招募到菌丝际中,作为营养物质刺激细菌的生长,影响菌丝际微生物群落结构[111]。例如,葡萄糖等菌丝分泌物刺激溶磷菌增殖,细菌沿着丛枝菌根真菌的菌丝移动,其中水膜和碳源都是细菌沿着真菌菌丝移动的必要条件[16]。豆科植物根际的丝状真菌所形成的菌丝网络可以作为根瘤菌运动的载体,促进根瘤菌与豆科植物接触,并影响根瘤的形成[112]。菌根是土壤环境中普遍存在的真菌-植物共生现象,可以作为微生物的栖息场所[113-116]。丛枝菌根真菌(arbuscular mycorrhizal fungi, AMF)与植物宿主形成共生关系的过程中,真菌菌丝首先穿过根表皮,继而侵入皮层细胞[116],这个过程为细菌进入根系细胞提供渠道。因此,我们推测,ARB在菌丝和菌根表面定殖后,会在根系分泌物、菌丝分泌物以及土壤理化性质的作用下发生移动,菌丝和菌根表面的水膜和营养物质可以提高ARB的移动能力,增加ARB接触根系的机会。随后,ARB可以借助菌丝和菌根进入根系细胞间隙或细胞内部,促进ARGs在植物中的进一步传递。同时,真菌-细菌生物膜以及菌根菌丝网络增加了细胞之间的接触频率,有助于ARGs在细菌与细菌、细菌与真菌以及细菌与植物之间的水平转移[17]。
3.3 污染物与环境胁迫重塑土壤-植物中ARGs组成
土壤是抗生素、重金属、杀生剂、微塑料以及人工纳米材料等污染物的重要储存库,影响抗生素抗性在土壤-植物中的传播[13, 117-119](图2(c))。重金属Cd胁迫下,通过调控土壤真菌群落促进了非根际土和根际土中ARGs丰度,同时促进了青葱内生菌中ARGs丰度,以及ARGs从植物地下部向地上部的转移[120]。施用直径~20 nm和~50 nm的Ag纳米材料以及Ag离子显著增加了土壤中外排泵基因的相对丰度,并通过抗生素和重金属抗性基因共选择机制改变了土壤抗生素抗性组成[121]。污染物可以直接影响植物根系[122],例如,可降解微塑料显著增加了菜豆根长和根瘤数目[123];小麦在Cd胁迫下,根系分泌的糖类随Cd浓度增加而增加,氨基酸随Cd浓度先增加后降低[124]。植物根部形态结构与根系分泌物的改变可以间接调控ARGs从土壤向植物的传播。此外,环境胁迫也会影响ARGs传播。Tan等[125]发现在盐渍土中,盐度从土壤深层向土壤表层逐渐增加,ARGs的水平转移频率随之降低,携带有抗性质粒的微生物的适应性也不断降低。高盐度促进了抗性质粒的去除,抑制了质粒的水平转移,因此可以降低表层土中ARGs从土壤向植物的传播。
3.4 土地利用方式与气候条件改变土壤-植物中ARGs组成
土地利用方式与气候条件也是植物抗性基因组演变的重要驱动因素(图2(c))。Xiang等[126]对比分析了城郊农田和森林地区的植物与土壤样品中ARGs和细菌群落,发现在受人类活动干扰更大的农田样品中,ARGs的组成更丰富、潜在ARGs宿主菌(厚壁菌门)丰度更高。Wang等[127]调查了降雨前、降雨期间、降雨后的土壤和雨水中ARGs的丰度,发现ARGs和MGEs在雨水中非常丰富,这表明雨水是一种独特的抗性基因库,降雨增加了土壤中ARGs丰度并且促进了土壤细菌的抵抗力,加速了ARGs和ARB的跨区域传播。ARGs和ARB随雨水进入土壤或与植物表面接触后,会进一步在土壤-植物系统中传播。
4 阻控抗生素抗性在土壤-植物中传播的技术途径(The control techniques to block the transmission of ARGs in the soil-plant system)
4.1 物理化学方法
好氧堆肥可以有效降低抗生素和ARGs丰度[128],从而在源头上阻控ARGs在农田环境中的传播扩散。好氧堆肥对ARGs的去除率可以达到25.4%~76.0%[128-129],施用堆肥后的牛粪,在小萝卜中的ARGs相对丰度明显降低[130]。Zhou等[131]开发的蒸汽爆破好氧堆肥(SEA-CBS)系统,对抗生素发酵残渣中抗生素去除率高达99.9%,极大降低了抗生素残留对土壤微生物抗性的选择压力。添加土壤改良剂在控制抗生素抗性传播方面也发挥重要作用。生物炭是一种常用的土壤改良剂,具有疏松多孔结构以及大表面积,为土壤微生物繁殖提供了充足的空间,而且通常具有丰富的化学基团可以吸附、水解、氧化和催化降解抗生素等化学物质[132]。因此添加生物炭可以提高土壤微生物多样性进而降低耐药菌优势,同时降低抗生素对土壤微生物抗性的选择压力。近年来,纳米材料逐渐被应用到去除抗生素抗性中,由于其特殊的尺寸结构,Fe2O3@MoS2纳米材料可以在细胞外大量累积,阻碍细胞间的物理接触,同时参与基因调控,降低质粒复制能力以及细胞膜通透性,降低ARGs的接合转移频率[133]。纳米级生物炭吸附胞外ARGs后不仅抑制了ARGs的复制,还通过羟基自由基破坏ARGs的双链结构,促进了ARGs的分解[134]。此外,纳米材料具有抗菌性能,可直接损伤细胞壁/细胞膜、引起氧化应激以及损害细胞内成分等[135],有望用于土壤-植物中ARB的去除。以上物理化学方法在阻断抗生素抗性传播方面具有良好的发展前景,然而这些技术也存在去除ARGs效果不稳定、存在潜在环境风险等隐患,例如,好氧堆肥对ARGs的去除并不彻底、生物炭施加到土壤中可能增加某些ARGs丰度[136]和纳米材料长期累积对土壤生物产生一定的毒性效应[137]等,这与技术本身以及实际施加的农田环境密切相关。因此,在开发阻断ARGs传播技术的同时,需要通过大量农田实验进行验证和推广,并加强环境风险的全面评估。
4.2 生物控制技术
生物控制技术借助土壤动物、植物和噬菌体,通过降解抗生素、改善微生物群落结构以及靶向灭活耐药病原菌等途径,可以有效控制ARGs在土壤-植物中的传播,近年来受到广泛关注。腐食动物蚯蚓可以通过中和土壤pH值和消耗有机物来促进四环素降解[138],降低城市污泥中喹诺酮类药物抗性基因的绝对丰度[139]。黑水虻幼虫、家蝇幼虫和蜣螂幼虫等食腐动物也对ARGs或抗生素有良好的去除效果[140-142]。此外,植物修复也被用于土壤环境抗性基因的阻控中[143]。研究发现,黑麦草(Lolium multiflorum L.)和芥菜(Brassica juncea L.)减少了土壤中的抗生素和抗性基因丰度[144]。Lu等[145]将满江红(Azolla imbricata)用作土壤改良剂,发现其可以促进抗生素的降解并降低有效态重金属的浓度、减少转座酶基因丰度,从而降低土壤中ARGs的丰度和多样性。植物抗菌素[146]作为抗生素替代物,可以从源头上缓解ARGs的累积与传播。龙脑香科、葡萄科等植物产生的白藜芦醇,作为具有生物活性的植物抗菌素,可以代替抗生素作为生物防治剂抑制病原体毒力[147],从而降低抗生素对微生物抗性的选择。噬菌体作为侵袭细菌的病毒,可用于控制土壤-植物中抗性微生物的传播扩散。Zhao等[21]研究了噬菌体疗法控制环境中抗生素抗性病原菌污染,结果表明噬菌体疗法能够有效地靶向灭活土壤-植物系统中的抗生素抗性病原菌,其中鸡尾酒噬菌体的效果最显著,多价噬菌体次之。值得注意的是,噬菌体也是细菌间交换遗传物质的重要载体,有可能促进ARGs在细菌之间的水平转移,因此要密切关注噬菌体等生物控制技术在实际土壤-植物系统中诱导抗生素抗性传播的潜在风险。
5 展望(Prospect)
抗生素抗性是当前全球关注的公共卫生与健康问题,ARGs随微生物从土壤转移到蔬菜作物,进入食物链,给人类带来潜在健康风险。目前研究虽然证实了ARGs从土壤迁移到植物的传播途径,但ARGs的具体迁移过程、传播机制尚不清楚。此外,为有效控制ARGs在土壤-植物中的传播,还需要结合其传播特性,采取合适的阻控手段。基于此,本文提出以下4个方面,值得在未来进一步关注。
(1)以微生物为载体的ARGs可以被植物吸收并在植物体内发生迁移,但在土壤环境中由于微生物代谢同时会产生大量的胞外ARGs。胞外ARGs是否会因为其较小的尺寸更容易被植物吸收、吸收后是否会在植物体内发生水平转移等,目前尚不清楚。
(2)ARB在植物内的形态与分布必然影响ARGs在植物中的进一步转移扩散,目前仍缺乏完善的技术手段对植物微生物组中的ARB和抗性组进行表征。
(3)已有研究证明了有机肥中残留的抗生素对土壤微生物进行抗性选择,但抗生素被植物吸收后,抗生素母体及其代谢产物是否会继续对内生菌进行压力选择,改变内生菌抗生素抗性?
(4)从源头上控制抗生素抗性的产生,是较为有效的阻控手段,但对于那些已经处于ARGs风险较高的农田生态系统,需要开发经济适用、环境友好的技术手段,结合ARGs在土壤-植物中的迁移特性,阻断其传播渠道。
通信作者简介:贾伟丽(1991—),女,博士,副研究员,主要研究方向为污染物环境行为与修复。
[1] Conde-Cid M, Nú ez-Delgado A, Fernández-Sanjurjo M, et al. Tetracycline and sulfonamide antibiotics in soils: Presence, fate and environmental risks [J]. Processes, 2020, 8(11): 1479
ez-Delgado A, Fernández-Sanjurjo M, et al. Tetracycline and sulfonamide antibiotics in soils: Presence, fate and environmental risks [J]. Processes, 2020, 8(11): 1479
[2] Yang Q L, Gao Y, Ke J, et al. Antibiotics: An overview on the environmental occurrence, toxicity, degradation, and removal methods [J]. Bioengineered, 2021, 12(1): 7376-7416
[3] Qiao M, Ying G G, Singer A C, et al. Review of antibiotic resistance in China and its environment [J]. Environment International, 2018, 110: 160-172
[4] 吴玉高, 李卓阳, 方菁. 畜禽粪便还田所致环境污染现况及其健康危害[J]. 环境与职业医学, 2021, 38(11): 1284-1290
Wu Y G, Li Z Y, Fang J. Environmental pollution and health hazards caused by animal agriculture manure [J]. Journal of Environmental and Occupational Medicine, 2021, 38(11): 1284-1290 (in Chinese)
[5] 朱冬, 陈青林, 丁晶, 等. 土壤生态系统中抗生素抗性基因与星球健康: 进展与展望[J]. 中国科学: 生命科学, 2019, 49(12): 1652-1663
Zhu D, Chen Q L, Ding J, et al. Antibiotic resistance genes in the soil ecosystem and planetary health: Progress and prospect [J]. Scientia Sinica (Vitae), 2019, 49(12): 1652-1663 (in Chinese)
[6] Ezzariai A, Hafidi M, Khadra A, et al. Human and veterinary antibiotics during composting of sludge or manure: Global perspectives on persistence, degradation, and resistance genes [J]. Journal of Hazardous Materials, 2018, 359: 465-481
[7] Qian X, Gu J, Sun W, et al. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting [J]. Journal of Hazardous Materials, 2018, 344: 716-722
[8] Xiang Q, Zhu D, Giles M, et al. Agricultural activities affect the pattern of the resistome within the phyllosphere microbiome in peri-urban environments [J]. Journal of Hazardous Materials, 2020, 382: 121068
[9] Pu Q, Wang H T, Pan T, et al. Enhanced removal of ciprofloxacin and reduction of antibiotic resistance genes by earthworm Metaphire vulgaris in soil [J]. The Science of the Total Environment, 2020, 742: 140409
[10] Zhu D, Delgado-Baquerizo M, Su J Q, et al. Deciphering potential roles of earthworms in mitigation of antibiotic resistance in the soils from diverse ecosystems [J]. Environmental Science &Technology, 2021, 55(11): 7445-7455
[11] Ding J, Zhu D, Hong B, et al. Long-term application of organic fertilization causes the accumulation of antibiotic resistome in earthworm gut microbiota [J]. Environment International, 2019, 124: 145-152
[12] Xu H, Chen Z Y, Huang R Y, et al. Antibiotic resistance gene-carrying plasmid spreads into the plant endophytic bacteria using soil bacteria as carriers [J]. Environmental Science &Technology, 2021, 55(15): 10462-10470
[13] 陈菲然, 许一诺, 杜昊, 等. 纳米材料与环境抗生素耐药性: 抗性基因流在土壤-植物系统中的迁移与阻断[J]. 科学通报, 2022, 67(35): 4206-4223
Chen F R, Xu Y N, Du H, et al. Nanomaterials and environmental antimicrobial resistance: Propagation and inhibition of antibiotic resistance gene flow in the soil-plant system [J]. Chinese Science Bulletin, 2022, 67(35): 4206-4223 (in Chinese)
[14] Müller D B, Vogel C, Bai Y, et al. The plant microbiota: Systems-level insights and perspectives [J]. Annual Review of Genetics, 2016, 50: 211-234
[15] Ba uelos-Vazquez L A, Torres Tejerizo G, Cervantes-De La Luz L, et al. Conjugative transfer between Rhizobium etli endosymbionts inside the root nodule [J]. Environmental Microbiology, 2019, 21(9): 3430-3441
uelos-Vazquez L A, Torres Tejerizo G, Cervantes-De La Luz L, et al. Conjugative transfer between Rhizobium etli endosymbionts inside the root nodule [J]. Environmental Microbiology, 2019, 21(9): 3430-3441
[16] Jiang F Y, Zhang L, Zhou J C, et al. Arbuscular mycorrhizal fungi enhance mineralisation of organic phosphorus by carrying bacteria along their extraradical hyphae [J]. The New Phytologist, 2021, 230(1): 304-315
[17] Ruan C J, Ramoneda J, Gogia G, et al. Fungal hyphae regulate bacterial diversity and plasmid-mediated functional novelty during range expansion [J]. Current Biology, 2022, 32(24): 5285-5294.e4
[18] Li S, Yao Q, Liu J J, et al. Liming mitigates the spread of antibiotic resistance genes in an acid black soil [J]. The Science of the Total Environment, 2022, 817: 152971
[19] Yuan W, Wang X J, Li Y, et al. Effects of struvite-loaded zeolite amendment on the fate of copper, tetracycline and antibiotic resistance genes in microplastic-contaminated soil [J]. Chemical Engineering Journal, 2022, 1: 430
[20] Zhang Y, Chen M L, Bao C X, et al. Application of pig manure compost with different biochar modifies the antibiotic resistome and bacterial community in agriculture soil [J]. Water Air and Soil Pollution, 2022, 233(4): 1
[21] Zhao Y C, Ye M, Zhang X T, et al. Comparing polyvalent bacteriophage and bacteriophage cocktails for controlling antibiotic-resistant bacteria in soil-plant system [J]. Science of the Total Environment, 2019, 657: 918-925
[22] 朱永官, 彭静静, 韦中, 等. 土壤微生物组与土壤健康[J]. 中国科学: 生命科学, 2021, 51(1): 1-11
Zhu Y G, Peng J J, Wei Z, et al. Linking the soil microbiome to soil health [J]. Scientia Sinica (Vitae), 2021, 51(1): 1-11 (in Chinese)
[23] 王娟, 王新华, 徐海. 多重耐药菌在人类、动物和环境的耐药和传播机制[J]. 微生物学报, 2016, 56(11): 1671-1679
Wang J, Wang X H, Xu H. Antimicrobial resistance and dissemination of multidrug resistant organisms—A review [J]. Acta Microbiologica Sinica, 2016, 56(11): 1671-1679 (in Chinese)
[24] Culp E J, Yim G, Waglechner N, et al. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics [J]. Nature Biotechnology, 2019, 37(10): 1149-1154
[25] 王顺, 王培红, 张楠, 等. 青霉素生产菌—产黄青霉的蛋白质组学研究进展[J]. 生物医学工程学杂志, 2015, 32(6): 1354-1358
Wang S, Wang P H, Zhang N, et al. Progress in proteomic study of the penicillin producer—Penicillium chrysogenum [J]. Journal of Biomedical Engineering, 2015, 32(6): 1354-1358 (in Chinese)
[26] Waksman S A. Streptomycin: Background, isolation, properties, and utilization [J]. Science, 1953, 118(3062): 259-266
[27] Peek J, Lilic M, Montiel D, et al. Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism [J]. Nature Communications, 2018, 9(1): 4147
[28] Nguyen B T, Chen Q L, He J Z, et al. Microbial regulation of natural antibiotic resistance: Understanding the protist-bacteria interactions for evolution of soil resistome [J]. The Science of the Total Environment, 2020, 705: 135882
[29] van Goethem M W, Pierneef R, Bezuidt O K I, et al. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils [J]. Microbiome, 2018, 6(1): 40
[30] Galán J C, González-Candelas F, Rolain J M, et al. Antibiotics as selectors and accelerators of diversity in the mechanisms of resistance: From the resistome to genetic plasticity in the β-lactamases world [J]. Frontiers in Microbiology, 2013, 4: 9
[31] Wang F, Fu Y H, Sheng H J, et al. Antibiotic resistance in the soil ecosystem: A One Health perspective [J]. Current Opinion in Environmental Science &Health, 2021, 20: 100230
[32] Pratama A A, van Elsas J D. The ‘neglected’ soil virome - potential role and impact [J]. Trends in Microbiology, 2018, 26(8): 649-662
[33] Calero-Cáceres W, Muniesa M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater [J]. Water Research, 2016, 95: 11-18
[34] Lee H S, Sobsey M D. Survival of prototype strains of somatic coliphage families in environmental waters and when exposed to UV low-pressure monochromatic radiation or heat [J]. Water Research, 2011, 45(12): 3723-3734
[35] Marti E, Jofre J, Balcazar J L. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant [J]. PLoS One, 2013, 8(10): e78906
[36] Srinivasiah S, Bhavsar J, Thapar K, et al. Phages across the biosphere: Contrasts of viruses in soil and aquatic environments [J]. Research in Microbiology, 2008, 159(5): 349-357
[37] Romanowski G, Lorenz M G, Sayler G, et al. Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay [J]. Applied and Environmental Microbiology, 1992, 58(9): 3012-3019
[38] Zhu B. Degradation of plasmid and plant DNA in water microcosms monitored by natural transformation and real-time polymerase chain reaction (PCR) [J]. Water Research, 2006, 40(17): 3231-3238
[39] 郑小伟, 黄力. 海洋病毒多样性及生态功能[J]. 中国科学基金, 2018, 32(5): 456-458
Zheng X W, Huang L. Diversity and ecological functions of marine viruses [J]. Bulletin of National Natural Science Foundation of China, 2018, 32(5): 456-458 (in Chinese)
[40] Larra aga O, Brown-Jaque M, Quirós P, et al. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil [J]. Environment International, 2018, 115: 133-141
aga O, Brown-Jaque M, Quirós P, et al. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil [J]. Environment International, 2018, 115: 133-141
[41] Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens [J]. Frontiers in Microbiology, 2018, 9: 2928
[42] Han X M, Hu H W, Chen Q L, et al. Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures [J]. Soil Biology and Biochemistry, 2018, 126: 91-102
[43] He L Y, Ying G G, Liu Y S, et al. Discharge of swine wastes risks water quality and food safety: Antibiotics and antibiotic resistance genes from swine sources to the receiving environments [J]. Environment International, 2016, 92-93: 210-219
[44] Liu W B, Ling N, Guo J J, et al. Dynamics of the antibiotic resistome in agricultural soils amended with different sources of animal manures over three consecutive years [J]. Journal of Hazardous Materials, 2021, 401: 123399
[45] Karkman A, Do T T, Walsh F, et al. Antibiotic-resistance genes in waste water [J]. Trends in Microbiology, 2018, 26(3): 220-228
[46] An X L, Su J Q, Li B, et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR [J]. Environment International, 2018, 117: 146-153
[47] Arun S, Kumar R M, Ruppa J, et al. Occurrence, sources and risk assessment of fluoroquinolones in dumpsite soil and sewage sludge from Chennai, India [J]. Environmental Toxicology and Pharmacology, 2020, 79: 103410
[48] Kyriacou A, Mitsou E, Abeliotis K, et al. Mapping of antibiotic resistant enterococci in wastewater treatment plants in Greece [J]. Desalination and Water Treatment, 2018, 112: 250-257
[49] Magee H Y, Maurer M M, Cobos A, et al. U.S. nationwide reconnaissance of ten infrequently monitored antibiotics in municipal biosolids [J]. The Science of the Total Environment, 2018, 643: 460-467
[50] Markowicz A, Bondarczuk K, Wiekiera A, et al. Is sewage sludge a valuable fertilizer? A soil microbiome and resistome study under field conditions [J]. Journal of Soils and Sediments, 2021, 21(8): 2882-2895
[51] Urra J, Alkorta I, Mijangos I, et al. Application of sewage sludge to agricultural soil increases the abundance of antibiotic resistance genes without altering the composition of prokaryotic communities [J]. Science of the Total Environment, 2019, 647: 1410-1420
[52] Zhu D, Ding J, Wang Y F, et al. Effects of trophic level and land use on the variation of animal antibiotic resistome in the soil food web [J]. Environmental Science &Technology, 2022, 56(21): 14937-14947
[53] König H. Bacillus species in the intestine of termites and other soil invertebrates [J]. Journal of Applied Microbiology, 2006, 101(3): 620-627
[54] Rea M C, Sit C S, Clayton E, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(20): 9352-9357
[55] Agamennone V, Roelofs D, van Straalen N M, et al. Antimicrobial activity in culturable gut microbial communities of springtails [J]. Journal of Applied Microbiology, 2018, 125(3): 740-752
[56] Zheng F, Zhu D, Giles M, et al. Mineral and organic fertilization alters the microbiome of a soil nematode Dorylaimus stagnalis and its resistome [J]. The Science of the Total Environment, 2019, 680: 70-78
[57] Zhu D, Xiang Q, Yang X R, et al. Trophic transfer of antibiotic resistance genes in a soil detritus food chain [J]. Environmental Science &Technology, 2019, 53(13): 7770-7781
[58] Zhu D, Ding J, Yin Y, et al. Effects of earthworms on the microbiomes and antibiotic resistomes of detritus fauna and phyllospheres [J]. Environmental Science &Technology, 2020, 54(10): 6000-6008
[59] Zheng F, Bi Q F, Giles M, et al. Fates of antibiotic resistance genes in the gut microbiome from different soil fauna under long-term fertilization [J]. Environmental Science &Technology, 2021, 55(1): 423-432
[60] Xiao Z, Han R, Su J, et al. Application of earthworm and silicon can alleviate antibiotic resistance in soil-Chinese cabbage system with ARGs contamination [J]. Environmental Pollution, 2023, 319: 120900
[61] Zhu D, Delgado-Baquerizo M, Su J Q, et al. Deciphering potential roles of earthworms in mitigation of antibiotic resistance in the soils from diverse ecosystems [J]. Environmental Science &Technology, 2021, 55(11): 7445-7455
[62] Wang C, Hu R W, Strong P J, et al. Prevalence of antibiotic resistance genes and bacterial pathogens along the soil-mangrove root continuum [J]. Journal of Hazardous Materials, 2021, 408: 124985
![]() O, Heuer H. Plant-nematode interactions assisted by microbes in the rhizosphere [J]. Current Issues in Molecular Biology, 2019, 30: 75-88
O, Heuer H. Plant-nematode interactions assisted by microbes in the rhizosphere [J]. Current Issues in Molecular Biology, 2019, 30: 75-88
[64] Fitzpatrick C R, Copeland J, Wang P W, et al. Assembly and ecological function of the root microbiome across angiosperm plant species [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(6): E1157-E1165
[65] Canarini A, Kaiser C, Merchant A, et al. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli [J]. Frontiers in Plant Science, 2019, 10: 157
[66] 李春霞, 吴兴彪, 靳亚忠. 根系代谢物介导的植物-微生物互作的研究进展[J]. 微生物学报, 2022, 62(9): 3318-3328
Li C X, Wu X B, Jin Y Z. Advances on plant-microbe interaction mediated by root metabolites [J]. Acta Microbiologica Sinica, 2022, 62(9): 3318-3328 (in Chinese)
[67] 周益帆, 白寅霜, 岳童, 等. 植物根际促生菌促生特性研究进展[J]. 微生物学通报, 2023, 50(2): 644-666
Zhou Y F, Bai Y S, Yue T, et al. Research progress on the growth-promoting characteristics of plant growth-promoting rhizobacteria [J]. Microbiology China, 2023, 50(2): 644-666 (in Chinese)
[68] Yu Y T, Zhang Q, Zhang Z Y, et al. Plants select antibiotic resistome in rhizosphere in early stage [J]. The Science of the Total Environment, 2023, 858(Pt 1): 159847
[69] Brandt K K, Sjøholm O R, Krogh K A, et al. Increased pollution-induced bacterial community tolerance to sulfadiazine in soil hotspots amended with artificial root exudates [J]. Environmental Science &Technology, 2009, 43(8): 2963-2968
[70] Yang Y, Zhang H, Fang Y, et al. Interference by non-host plant roots and root exudates in the infection processes of Phytophthora nicotianae [J]. Frontiers of Agricultural Science and Engineering, 2021, 8(3): 447-459
[71] Herms C H, Hennessy R C, Bak F, et al. Back to our roots: Exploring the role of root morphology as a mediator of beneficial plant-microbe interactions [J]. Environmental Microbiology, 2022, 24(8): 3264-3272
[72] Hijwegen T. Lignification, a possible mechanism of active resistance against pathogens [J]. Netherlands Journal of Plant Pathology, 1963, 69(6): 314-317
[73] Barberon M, Vermeer J E M, De Bellis D, et al. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation [J]. Cell, 2016, 164(3): 447-459
[74] Wang P, Wang C M, Gao L, et al. Aliphatic suberin confers salt tolerance to Arabidopsis by limiting Na+ influx, K+ efflux and water backflow [J]. Plant and Soil, 2020, 448: 603-620
[75] Yue L, Chen F R, Yu K Q, et al. Early development of apoplastic barriers and molecular mechanisms in juvenile maize roots in response to La2O3 nanoparticles [J]. The Science of the Total Environment, 2019, 653: 675-683
[76] Ku Y S, Wang Z L, Duan S W, et al. Rhizospheric communication through mobile genetic element transfers for the regulation of microbe-plant interactions [J]. Biology, 2021, 10(6): 477
[77] Forsberg K J, Patel S, Gibson M K, et al. Bacterial phylogeny structures soil resistomes across habitats [J]. Nature, 2014, 509(7502): 612-616
[78] Cui E P, Gao F, Liu Y, et al. Amendment soil with biochar to control antibiotic resistance genes under unconventional water resources irrigation: Proceed with caution [J]. Environmental Pollution, 2018, 240: 475-484
[79] Tan S Y, Yang C L, Mei X L, et al. The effect of organic acids from tomato root exudates on rhizosphere colonization of Bacillus amyloliquefaciens T-5 [J]. Applied Soil Ecology, 2013, 64: 15-22
[80] Wang Y J, Kou S M, Jiang Q Y, et al. Factors affecting transfer of degradative plasmids between bacteria in soils [J]. Applied Soil Ecology, 2014, 84: 254-261
[81] Mølbak L, Molin S, Kroer N. Root growth and exudate production define the frequency of horizontal plasmid transfer in the rhizosphere [J]. FEMS Microbiology Ecology, 2007, 59(1): 167-176
[82] 曾庆涛. 农田土壤典型抗生素抗性基因污染及其土——气迁移研究[D]. 杭州: 浙江大学, 2019: 8-11
Zeng Q T. Study on typical antibiotic resistance gene pollution and soil-air migration in farmland soil [D].Hangzhou: Zhejiang University, 2019: 8-11 (in Chinese)
[83] Deng B Q, Li W, Lu H J, et al. Film mulching reduces antibiotic resistance genes in the phyllosphere of lettuce [J]. Journal of Environmental Sciences (China), 2022, 112: 121-128
[84] Han X M, Hu H W, Li J Y, et al. Long-term application of swine manure and sewage sludge differently impacts antibiotic resistance genes in soil and phyllosphere [J]. Geoderma, 2022, 1: 411
[85] Melotto M, Zhang L, Oblessuc P R, et al. Stomatal defense a decade later [J]. Plant Physiology, 2017, 174(2): 561-571
[86] Ruppel S, Krumbein A, Schreiner M. Composition of the phyllospheric microbial populations on vegetable plants with different glucosinolate and carotenoid compositions [J]. Microbial Ecology, 2008, 56(2): 364-372
[87] Farré-Armengol G, Filella I, Llusia J, et al. Bidirectional interaction between phyllospheric microbiotas and plant volatile emissions [J]. Trends in Plant Science, 2016, 21(10): 854-860
[88] 杨宽, 王慧玲, 叶坤浩, 等. 叶际微生物及与植物互作的研究进展[J]. 云南农业大学学报(自然科学), 2021, 36(1): 155-164
Yang K, Wang H L, Ye K H, et al. Advances in research on phyllosphere microorganisms and their interaction with plants [J]. Journal of Yunnan Agricultural University (Natural Science), 2021, 36(1): 155-164 (in Chinese)
[89] Chen Q L, Cui H L, Su J Q, et al. Antibiotic resistomes in plant microbiomes [J]. Trends in Plant Science, 2019, 24(6): 530-541
[90] Zhang Y J, Hu H W, Chen Q L, et al. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes [J]. Environment International, 2019, 130: 104912
[91] Zhu B K, Chen Q L, Chen S C, et al. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? [J]. Environment International, 2017, 98: 152-159
[92] Pu Q, Zhao L X, Li Y T, et al. Manure fertilization increase antibiotic resistance in soils from typical greenhouse vegetable production bases, China [J]. Journal of Hazardous Materials, 2020, 391: 122267
[93] Zhang Y J, Hu H W, Chen Q L, et al. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes [J]. Environment International, 2019, 130: 104912
[94] 易婷, 缪煜轩, 冯永君. 内生菌与植物的相互作用: 促生与生物薄膜的形成[J]. 微生物学通报, 2008, 35(11): 1774-1780
Yi T, Miao Y X, Feng Y J. Plant-endophyte interaction: Growth-promoting effect of endophytes and their biofilm formation [J]. Microbiology, 2008, 35(11): 1774-1780 (in Chinese)
[95] Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure [J]. Current Opinion in Biotechnology, 2003, 14(3): 255-261
[96] 李港, 刘博文, 王子晗, 等. 植物内生菌研究进展[J]. 现代农业科技, 2022(17): 110-113
Li G, Liu B W, Wang Z H, et al. Research progress on plant endophytes [J]. Modern Agricultural Science and Technology, 2022(17): 110-113 (in Chinese)
[97] Jiao W T, Du R J, Ye M, et al. ‘Agricultural Waste to Treasure’ - Biochar and eggshell to impede soil antibiotics/antibiotic resistant bacteria (genes) from accumulating in Solanum tuberosum L. [J]. Environmental Pollution, 2018, 242(Pt B): 2088-2095
[98] Zhang W G, Zhang M, Li W. The antibiotic resistance genes contamination of strawberries with the long-term use of raw, aerobic composting, and anaerobic composting livestock manure: A comparative study [J]. Frontiers in Environmental Science, 2022, 1: 721
[99] Wang Y Z, Zhou S Y, Zhou X Y, et al. Manure and biochar have limited effect on lettuce leaf endophyte resistome [J]. The Science of the Total Environment, 2023, 860: 160515
[100] Sanz C, Casadoi M, Tadic -D, et al. Impact of organic soil amendments in antibiotic levels, antibiotic resistance gene loads, and microbiome composition in corn fields and crops [J]. Environmental Research, 2022, 214(Pt 1): 113760
[101] Seyoum M M, Lichtenberg R, Orlofsky E, et al. Antibiotic resistance in soil and tomato crop irrigated with freshwater and two types of treated wastewater [J]. Environmental Research, 2022, 211: 113021
[102] Brambilla S, Frare R, Stritzler M, et al. Synthetic multi-antibiotic resistant plasmids in plant-associated bacteria from agricultural soils [J]. Journal of Global Antimicrobial Resistance, 2020, 22: 113-116
[103] Tzec-Gamboa M, Solorio-Sánchez F, Fiebrig I, et al. Biochemical and molecular characterization of native rhizobia nodulating Leucaena leucocephala with potential use as bioinoculants in Yucatan, Mexico [J]. Chiang Mai Journal of Science, 2020, 47(1): 1-15
[104] Qaddawi Z T, Mohammed A A-H. Microbacterium sp. AJ-Z. isolated from the root nodules of fenugreek (Trigonella foenum-graecum) [J]. Biochemical and Cellular Archives, 2021, 1: 4457-4560
[105] Liu B S, Zhang D Y, Pan X L. Nodules of wild legumes as unique natural hotspots of antibiotic resistance genes [J]. The Science of the Total Environment, 2022, 839: 156036
[106] Chou M X, Sun Y L, Yang J Y, et al. Comprehensive analysis of phenotype, microstructure and global transcriptional profiling to unravel the effect of excess copper on the symbiosis between nitrogen-fixing bacteria and Medicago lupulina [J]. Science of the Total Environment, 2019, 656: 1346-1357
[107] Rangel W M, de Oliveira Longatti S M, Ferreira P A A, et al. Leguminosae native nodulating bacteria from a gold mine As-contaminated soil: Multi-resistance to trace elements, and possible role in plant growth and mineral nutrition [J]. International Journal of Phytoremediation, 2017, 19(10): 925-936
[108] Marchou B, Bellido F, Charnas R, et al. Contribution of beta-lactamase hydrolysis and outer membrane permeability to ceftriaxone resistance in Enterobacter cloacae [J]. Antimicrobial Agents and Chemotherapy, 1987, 31(10): 1589-1595
[109] Baker-Austin C, Wright M S, Stepanauskas R, et al. Co-selection of antibiotic and metal resistance [J]. Trends in Microbiology, 2006, 14(4): 176-182
[110] Ren Z, Jeckel H, Simon-Soro A, et al. InterKingdom assemblages in human saliva display group-level surface mobility and disease-promoting emergent functions [J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(41): e2209699119
[111] Zhang L, Zhou J C, George T S, et al. Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra [J]. Trends in Plant Science, 2022, 27(4): 402-411
[112] Zhang W, Li X G, Sun K, et al. Mycelial network-mediated rhizobial dispersal enhances legume nodulation [J]. The ISME Journal, 2020, 14(4): 1015-1029
[113] 金文豪, 邵帅, 陈俊辉, 等. 不同类型菌根对土壤碳循环的影响差异研究进展[J]. 浙江农林大学学报, 2021, 38(5): 953-962
Jin W H, Shao S, Chen J H, et al. Research progress in the impact of different mycorrhizal types on soil carbon cycling [J]. Journal of Zhejiang A &F University, 2021, 38(5): 953-962 (in Chinese)
[114] Liang M X, Johnson D, Burslem D F R P, et al. Soil fungal networks maintain local dominance of ectomycorrhizal trees [J]. Nature Communications, 2020, 11(1): 2636
[115] Chen L, Swenson N G, Ji N N, et al. Differential soil fungus accumulation and density dependence of trees in a subtropical forest [J]. Science, 2019, 366(6461): 124-128
[116] 曹本福, 姜海霞, 刘丽, 等. 丛枝菌根菌丝网络在植物互作中的作用机制研究进展[J]. 应用生态学报, 2021, 32(9): 3385-3396
Cao B F, Jiang H X, Liu L, et al. Research progress on mechanism of arbuscular common mycorrhizal networks in plant-plant interactions [J]. Chinese Journal of Applied Ecology, 2021, 32(9): 3385-3396 (in Chinese)
[117] 赵方杰, 谢婉滢, 汪鹏. 土壤与人体健康[J]. 土壤学报, 2020, 57(1): 1-11
Zhao F J, Xie W Y, Wang P. Soil and human health [J]. Acta Pedologica Sinica, 2020, 57(1): 1-11 (in Chinese)
[118] Qi L, Ge Y, Xia T, et al. Rare earth oxide nanoparticles promote soil microbial antibiotic resistance by selectively enriching antibiotic resistance genes [J]. Environmental Science: Nano, 2019, 6(2): 456-466
[119] Hu H W, Wang J T, Li J, et al. Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils [J]. Environmental Microbiology, 2016, 18(11): 3896-3909
[120] Wei H W, Ding S, Qiao Z R, et al. Insights into factors driving the transmission of antibiotic resistance from sludge compost-amended soil to vegetables under cadmium stress [J]. The Science of the Total Environment, 2020, 729: 138990
[121] Chen Q L, Zhu D, An X L, et al. Does nano silver promote the selection of antibiotic resistance genes in soil and plant? [J]. Environment International, 2019, 128: 399-406
[122] Chen G L, Li Y Z, Liu S L, et al. Effects of micro(nano)plastics on higher plants and the rhizosphere environment [J]. The Science of the Total Environment, 2022, 807(Pt 1): 150841
[123] Meng F R, Yang X M, Riksen M, et al. Response of common bean (Phaseolus vulgaris L.) growth to soil contaminated with microplastics [J]. The Science of the Total Environment, 2021, 755(Pt 2): 142516
[124] 张玲, 王焕校. 镉胁迫下小麦根系分泌物的变化[J]. 生态学报, 2002, 22(4): 496-502
Zhang L, Wang H X. Changes of root exudates to cadmium stress in wheat (Triticum aestivm L.) [J]. Acta Ecologica Sinica, 2002, 22(4): 496-502 (in Chinese)
[125] Tan L, Wang F, Liang M M, et al. Antibiotic resistance genes attenuated with salt accumulation in saline soil [J]. Journal of Hazardous Materials, 2019, 374: 35-42
[126] Xiang Q, Zhu D, Giles M, et al. Agricultural activities affect the pattern of the resistome within the phyllosphere microbiome in peri-urban environments [J]. Journal of Hazardous Materials, 2020, 382: 121068
[127] Wang Q, Guo S Y, Hou Z L, et al. Rainfall facilitates the transmission and proliferation of antibiotic resistance genes from ambient air to soil [J]. The Science of the Total Environment, 2021, 799: 149260
[128] Zhang M, He L Y, Liu Y S, et al. Variation of antibiotic resistome during commercial livestock manure composting [J]. Environment International, 2020, 136: 105458
[129] Liu B T, Yu K F, Ahmed I, et al. Key factors driving the fate of antibiotic resistance genes and controlling strategies during aerobic composting of animal manure: A review [J]. The Science of the Total Environment, 2021, 791: 148372
[130] Guron G K P, Arango-Argoty G, Zhang L Q, et al. Effects of dairy manure-based amendments and soil texture on lettuce- and radish-associated microbiota and resistomes [J]. mSphere, 2019, 4(3): e00239-e00219
[131] Zhou J Y, Ping R, Wu H, et al. Recycling of neomycin fermentation residue using SEA-CBS technology: Growth performance and antibiotic resistance genes [J]. The Science of the Total Environment, 2022, 807(Pt 1): 150860
[132] Zhao Z D, Zhou W J. Insight into interaction between biochar and soil minerals in changing biochar properties and adsorption capacities for sulfamethoxazole [J]. Environmental Pollution, 2019, 245: 208-217
[133] Wang H G, Qi H C, Zhu M, et al. MoS2 decorated nanocomposite: Fe2O3@MoS2 inhibits the conjugative transfer of antibiotic resistance genes [J]. Ecotoxicology and Environmental Safety, 2019, 186: 109781
[134] Lian F, Yu W C, Zhou Q X, et al. Size matters: Nano-biochar triggers decomposition and transformation inhibition of antibiotic resistance genes in aqueous environments [J]. Environmental Science &Technology, 2020, 54(14): 8821-8829
[135] Makabenta J M V, Nabawy A, Li C H, et al. Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections [J]. Nature Reviews Microbiology, 2021, 19(1): 23-36
[136] 汤冬梅, 武俊梅, 黄永炳, 等. 生物炭添加对土壤中抗生素和抗性基因的环境行为影响研究进展[J]. 环境化学, 2022, 41(6): 1957-1966
Tang D M, Wu J M, Huang Y B, et al. Research advances in the effect of biochar amendment on environmental behaviors of antibiotics and antibiotic resistance genes in soils [J]. Environmental Chemistry, 2022, 41(6): 1957-1966 (in Chinese)
[137] Dinesh R, Anandaraj M, Srinivasan V, et al. Engineered nanoparticles in the soil and their potential implications to microbial activity [J]. Geoderma, 2012, 173-174: 19-27
[138] Lin Z, Zhen Z, Luo S W, et al. Effects of two ecological earthworm species on tetracycline degradation performance, pathway and bacterial community structure in laterite soil [J]. Journal of Hazardous Materials, 2021, 412: 125212
[139] Cui G Y, Li F S, Li S L, et al. Changes of quinolone resistance genes and their relations with microbial profiles during vermicomposting of municipal excess sludge [J]. The Science of the Total Environment, 2018, 644: 494-502
[140] Liu C C, Yao H Y, Chapman S J, et al. Changes in gut bacterial communities and the incidence of antibiotic resistance genes during degradation of antibiotics by black soldier fly larvae [J]. Environment International, 2020, 142: 105834
[141] Wang H, Li H Y, Gilbert J A, et al. Housefly larva vermicomposting efficiently attenuates antibiotic resistance genes in swine manure, with concomitant bacterial population changes [J]. Applied and Environmental Microbiology, 2015, 81(22): 7668-7679
[142] Zhao X, Shen J P, Shu C L, et al. Attenuation of antibiotic resistance genes in livestock manure through vermicomposting via Protaetia brevitarsis and its fate in a soil-vegetable system [J]. The Science of the Total Environment, 2022, 807(Pt 1): 150781
[143] Liang Y T, Pei M, Wang D D, et al. Improvement of soil ecosystem multifunctionality by dissipating manure-induced antibiotics and resistance genes [J]. Environmental Science &Technology, 2017, 51(9): 4988-4998
[144] Cui E P, Cui B J, Fan X Y, et al. Ryegrass (Lolium multiflorum L.) and Indian mustard (Brassica juncea L.) intercropping can improve the phytoremediation of antibiotics and antibiotic resistance genes but not heavy metals [J]. The Science of the Total Environment, 2021, 784: 147093
[145] Lu X M, Lu P Z. Distribution of antibiotic resistance genes in soil amended using Azolla imbricata and its driving mechanisms [J]. The Science of the Total Environment, 2019, 692: 422-431
[146] Zhang Q, Zhang Z Y, Zhou S, et al. Macleaya cordata extract, an antibiotic alternative, does not contribute to antibiotic resistance gene dissemination [J]. Journal of Hazardous Materials, 2021, 412: 125272
[147] Kang J E, Yoo N, Jeon B J, et al. Resveratrol oligomers, plant-produced natural products with anti-virulence and plant immune-priming roles [J]. Frontiers in Plant Science, 2022, 13: 885625